Overview
Automated methods to detect and classify human diseases from medical images.
Data Description
The dataset is organized into 3 folders (train, test, val) and contains subfolders for each image category (Pneumonia/Normal). There are 5,863 X-Ray images (JPEG) and 2 categories (Pneumonia/Normal).
Chest X-ray images (anterior-posterior) were selected from retrospective cohorts of pediatric patients of one to five years old from Guangzhou Women and Children’s Medical Center, Guangzhou. All chest X-ray imaging was performed as part of patients’ routine clinical care.
For the analysis of chest x-ray images, all chest radiographs were initially screened for quality control by removing all low quality or unreadable scans. The diagnoses for the images were then graded by two expert physicians before being cleared for training the AI system. In order to account for any grading errors, the evaluation set was also checked by a third expert.
You can find the dataset here.
Files
test
- NORMAL
- PNEUMONIA
train
- NORMAL
- PNEUMONIA
val
- NORMAL
- PNEUMONIA
So let’s begin here…
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from PIL import Image
from keras.models import Sequential
from keras.layers import Conv2D
from keras.layers import MaxPooling2D
from keras.layers import Flatten
from keras.layers import Dense
from keras.preprocessing.image import ImageDataGenerator, load_img
import os
Load Data
train_folder= '../input/chest-xray-pneumonia/chest_xray/train/'
val_folder = '../input/chest-xray-pneumonia/chest_xray/val/'
test_folder = '../input/chest-xray-pneumonia/chest_xray/test/'
Train Data
train_n = train_folder+'NORMAL/'
train_p = train_folder+'PNEUMONIA/'
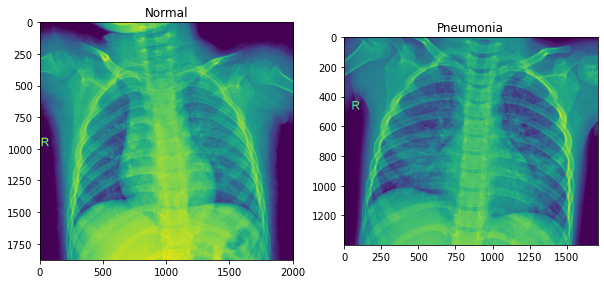
Let’s have a look at our data
#Normal pic
rand_norm = np.random.randint(0,len(os.listdir(train_n)))
norm_pic = os.listdir(train_n)[rand_norm]
norm_pic_address = train_n + norm_pic
#Pneumonia
rand_p = np.random.randint(0,len(os.listdir(train_p)))
pne_pic = os.listdir(train_p)[rand_p]
pne_pic_address = train_p + pne_pic
#Let's plot these images
f = plt.figure(figsize= (10,6))
a1 = f.add_subplot(1,2,1)
img_plot = plt.imshow(Image.open(norm_pic_address))
a1.set_title('Normal')
a2 = f.add_subplot(1, 2, 2)
img_plot = plt.imshow(Image.open(pne_pic_address))
a2.set_title('Pneumonia')
Defining Model
cnn_model = Sequential()
cnn_model.add(Conv2D(32, (3, 3), activation="relu", input_shape=(64, 64, 3)))
cnn_model.add(MaxPooling2D(pool_size = (2, 2)))
cnn_model.add(Conv2D(32, (3, 3), activation="relu"))
cnn_model.add(MaxPooling2D(pool_size = (2, 2)))
cnn_model.add(Flatten())
cnn_model.add(Dense(activation = 'relu', units = 128))
cnn_model.add(Dense(activation = 'sigmoid', units = 1))
Compile Model
cnn_model.compile(optimizer = 'adam', loss = 'binary_crossentropy', metrics = ['accuracy'])
train_datagen = ImageDataGenerator(rescale = 1./255,
shear_range = 0.2,
zoom_range = 0.2,
horizontal_flip = True)
train_set = train_datagen.flow_from_directory(train_folder,
target_size = (64, 64),
batch_size = 32,
class_mode = 'binary')
Found 5216 images belonging to 2 classes.
test_datagen = ImageDataGenerator(rescale = 1./255)
validation_generator = test_datagen.flow_from_directory(val_folder,
target_size=(64, 64),
batch_size=32,
class_mode='binary')
test_set = test_datagen.flow_from_directory(test_folder,
target_size = (64, 64),
batch_size = 32,
class_mode = 'binary')
Found 16 images belonging to 2 classes.
Found 624 images belonging to 2 classes.
Fit Model
cnn_model_his = cnn_model.fit_generator(train_set,
steps_per_epoch = 163,
epochs = 10,
validation_data = validation_generator,
validation_steps = 624)
Epoch 1/10
163/163 [==============================] - 238s 1s/step - loss: 0.3814 - accuracy: 0.8351 - val_loss: 0.4382 - val_accuracy: 0.8125
Epoch 2/10
163/163 [==============================] - 211s 1s/step - loss: 0.2354 - accuracy: 0.9045 - val_loss: 0.5071 - val_accuracy: 0.7500
Epoch 3/10
163/163 [==============================] - 217s 1s/step - loss: 0.2123 - accuracy: 0.9160 - val_loss: 0.4330 - val_accuracy: 0.8125
Epoch 4/10
163/163 [==============================] - 213s 1s/step - loss: 0.1862 - accuracy: 0.9231 - val_loss: 0.3393 - val_accuracy: 0.7500
Epoch 5/10
163/163 [==============================] - 208s 1s/step - loss: 0.1732 - accuracy: 0.9302 - val_loss: 0.4586 - val_accuracy: 0.7500
Epoch 6/10
163/163 [==============================] - 209s 1s/step - loss: 0.1601 - accuracy: 0.9388 - val_loss: 0.4599 - val_accuracy: 0.6875
Epoch 7/10
163/163 [==============================] - 208s 1s/step - loss: 0.1600 - accuracy: 0.9344 - val_loss: 0.5183 - val_accuracy: 0.7500
Epoch 8/10
163/163 [==============================] - 213s 1s/step - loss: 0.1382 - accuracy: 0.9479 - val_loss: 0.3811 - val_accuracy: 0.8125
Epoch 9/10
163/163 [==============================] - 211s 1s/step - loss: 0.1524 - accuracy: 0.9411 - val_loss: 0.8907 - val_accuracy: 0.6250
Epoch 10/10
163/163 [==============================] - 211s 1s/step - loss: 0.1357 - accuracy: 0.9463 - val_loss: 0.3493 - val_accuracy: 0.8125
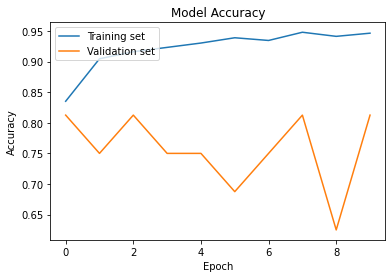
Evaluate Model
Accuracy
test_acc = cnn_model.evaluate_generator(test_set,steps=624)
print('The testing accuracy is :',test_acc[1]*100, '%')
The testing accuracy is : 91.36195778846741 %
plt.plot(cnn_model_his.history['accuracy'])
plt.plot(cnn_model_his.history['val_accuracy'])
plt.title('Model Accuracy')
plt.ylabel('Accuracy')
plt.xlabel('Epoch')
plt.legend(['Training set', 'Validation set'], loc='upper left')
plt.show()
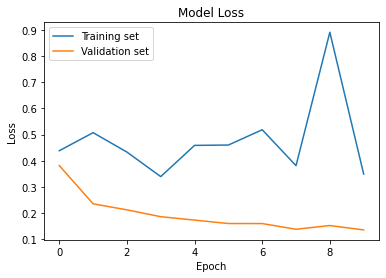
Loss
plt.plot(cnn_model_his.history['val_loss'])
plt.plot(cnn_model_his.history['loss'])
plt.title('Model Loss')
plt.ylabel('Loss')
plt.xlabel('Epoch')
plt.legend(['Training set', 'Validation set'], loc='upper left')
plt.show()