Content
The dataset contains 2 folders
- Infected
- Uninfected
And a total of 27,558 images.
Acknowledgements
This Dataset is taken from the official NIH Website: https://ceb.nlm.nih.gov/repositories/malaria-datasets/ And uploaded here, so anybody trying to start working with this dataset can get started immediately, as to download the dataset from NIH website is quite slow. Photo by Егор Камелев on Unsplash https://unsplash.com/@ekamelev
Inspiration
Save humans by detecting and deploying Image Cells that contain Malaria or not!
So let’s begin here…
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import keras
from keras import Sequential
from keras.layers import Conv2D,MaxPool2D,Dropout,Flatten,Dense,BatchNormalization
from keras.preprocessing.image import ImageDataGenerator
from PIL import Image
import os
print(os.listdir("../input/cell-images-for-detecting-malaria/cell_images"))
[‘Parasitized’, ‘Uninfected’, ‘cell_images’]
Set image size
width = 128
height = 128
Let’s have a look at our data
infected_folder = '../input/cell-images-for-detecting-malaria/cell_images/Parasitized/'
uninfected_folder = '../input/cell-images-for-detecting-malaria/cell_images/Uninfected/'
print(len(os.listdir(infected_folder)))
print(len(os.listdir(uninfected_folder)))
13780 13780
Let’s have a look at our data
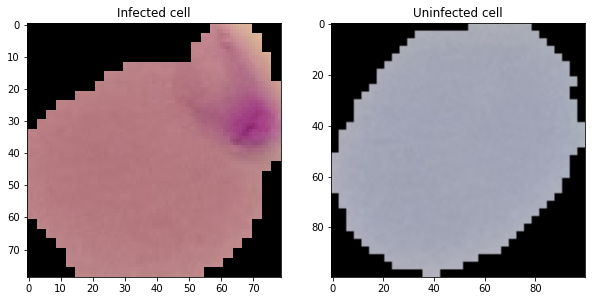
# Infected cell image
rand_inf = np.random.randint(0,len(os.listdir(infected_folder)))
inf_pic = os.listdir(infected_folder)[rand_inf]
#Uninfected cell image
rand_uninf = np.random.randint(0,len(os.listdir(uninfected_folder)))
uninf_pic = os.listdir(uninfected_folder)[rand_uninf]
# Load the images
inf_load = Image.open(infected_folder+inf_pic)
uninf_load = Image.open(uninfected_folder+uninf_pic)
# Let's plt these images
f = plt.figure(figsize= (10,6))
a1 = f.add_subplot(1,2,1)
img_plot = plt.imshow(inf_load)
a1.set_title('Infected cell')
a2 = f.add_subplot(1, 2, 2)
img_plot = plt.imshow(uninf_load)
a2.set_title('Uninfected cell')
Dividing data in train and test
datagen = ImageDataGenerator(rescale=1/255.0, validation_split=0.2)
Train Data
trainDatagen = datagen.flow_from_directory(directory='../input/cell-images-for-detecting-malaria/cell_images/cell_images/',
target_size=(width,height),
class_mode = 'binary',
batch_size = 16,
subset='training')
Found 22048 images belonging to 2 classes.
Validation Data
valDatagen = datagen.flow_from_directory(directory='../input/cell-images-for-detecting-malaria/cell_images/cell_images/',
target_size=(width,height),
class_mode = 'binary',
batch_size = 16,
subset='validation')
Found 5510 images belonging to 2 classes.
Create Model
We will create a CNN model where we will put 128x128 image with 3 channels(RGB) and will get a result as Infected or Uninfected.
model = Sequential()
model.add(Conv2D(16,(3,3),activation='relu',input_shape=(128,128,3)))
model.add(MaxPool2D(2,2))
model.add(Dropout(0.2))
model.add(Conv2D(32,(3,3),activation='relu'))
model.add(MaxPool2D(2,2))
model.add(Dropout(0.3))
model.add(Conv2D(64,(3,3),activation='relu'))
model.add(MaxPool2D(2,2))
model.add(Dropout(0.3))
model.add(Flatten())
model.add(Dense(64,activation='relu'))
model.add(Dropout(0.5))
model.add(Dense(1,activation='sigmoid'))
Compile Model
model.compile(optimizer='adam',loss='binary_crossentropy',metrics=['accuracy'])
Fit Model
cnn_model = model.fit_generator(generator = trainDatagen,
steps_per_epoch = len(trainDatagen),
epochs =20,
validation_data = valDatagen,
validation_steps=len(valDatagen))
Epoch 1/20
1378/1378 [==============================] - 107s 78ms/step - loss: 0.5049 - accuracy: 0.7332 - val_loss: 0.1742 - val_accuracy: 0.9334
Epoch 2/20
1378/1378 [==============================] - 46s 33ms/step - loss: 0.1827 - accuracy: 0.9427 - val_loss: 0.1584 - val_accuracy: 0.9448
Epoch 3/20
1378/1378 [==============================] - 47s 34ms/step - loss: 0.1616 - accuracy: 0.9514 - val_loss: 0.1635 - val_accuracy: 0.9417
Epoch 4/20
1378/1378 [==============================] - 46s 33ms/step - loss: 0.1477 - accuracy: 0.9554 - val_loss: 0.1662 - val_accuracy: 0.9434
Epoch 5/20
1378/1378 [==============================] - 47s 34ms/step - loss: 0.1388 - accuracy: 0.9572 - val_loss: 0.1682 - val_accuracy: 0.9483
Epoch 6/20
1378/1378 [==============================] - 47s 34ms/step - loss: 0.1326 - accuracy: 0.9590 - val_loss: 0.1872 - val_accuracy: 0.9417
Epoch 7/20
1378/1378 [==============================] - 48s 35ms/step - loss: 0.1314 - accuracy: 0.9583 - val_loss: 0.1830 - val_accuracy: 0.9475
Epoch 8/20
1378/1378 [==============================] - 46s 34ms/step - loss: 0.1305 - accuracy: 0.9597 - val_loss: 0.1670 - val_accuracy: 0.9474
Epoch 9/20
1378/1378 [==============================] - 47s 34ms/step - loss: 0.1236 - accuracy: 0.9589 - val_loss: 0.1799 - val_accuracy: 0.9470
Epoch 10/20
1378/1378 [==============================] - 49s 36ms/step - loss: 0.1219 - accuracy: 0.9614 - val_loss: 0.1635 - val_accuracy: 0.9439
Epoch 11/20
1378/1378 [==============================] - 50s 36ms/step - loss: 0.1189 - accuracy: 0.9610 - val_loss: 0.1697 - val_accuracy: 0.9423
Epoch 12/20
1378/1378 [==============================] - 47s 34ms/step - loss: 0.1184 - accuracy: 0.9601 - val_loss: 0.1735 - val_accuracy: 0.9465
Epoch 13/20
1378/1378 [==============================] - 44s 32ms/step - loss: 0.1161 - accuracy: 0.9613 - val_loss: 0.1694 - val_accuracy: 0.9461
Epoch 14/20
1378/1378 [==============================] - 44s 32ms/step - loss: 0.1138 - accuracy: 0.9628 - val_loss: 0.1716 - val_accuracy: 0.9430
Epoch 15/20
1378/1378 [==============================] - 45s 32ms/step - loss: 0.1126 - accuracy: 0.9628 - val_loss: 0.1640 - val_accuracy: 0.9468
Epoch 16/20
1378/1378 [==============================] - 45s 33ms/step - loss: 0.1153 - accuracy: 0.9623 - val_loss: 0.1991 - val_accuracy: 0.9432
Epoch 17/20
1378/1378 [==============================] - 44s 32ms/step - loss: 0.1083 - accuracy: 0.9648 - val_loss: 0.1704 - val_accuracy: 0.9456
Epoch 18/20
1378/1378 [==============================] - 87s 63ms/step - loss: 0.1072 - accuracy: 0.9630 - val_loss: 0.1772 - val_accuracy: 0.9466
Epoch 19/20
1378/1378 [==============================] - 71s 52ms/step - loss: 0.1076 - accuracy: 0.9643 - val_loss: 0.1638 - val_accuracy: 0.9452
Epoch 20/20
1378/1378 [==============================] - 47s 34ms/step - loss: 0.1068 - accuracy: 0.9641 - val_loss: 0.1833 - val_accuracy: 0.9459
Evaluation
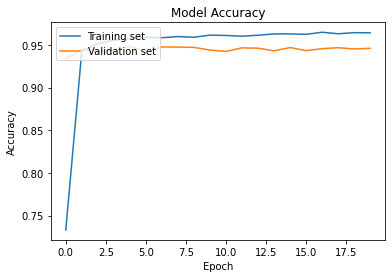
Accuracy
plt.plot(cnn_model.history['accuracy'])
plt.plot(cnn_model.history['val_accuracy'])
plt.title('Model Accuracy')
plt.ylabel('Accuracy')
plt.xlabel('Epoch')
plt.legend(['Training set', 'Validation set'], loc='upper left')
plt.show()
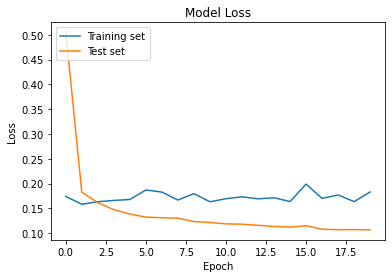
Loss
plt.plot(cnn_model.history['val_loss'])
plt.plot(cnn_model.history['loss'])
plt.title('Model Loss')
plt.ylabel('Loss')
plt.xlabel('Epoch')
plt.legend(['Training set', 'Test set'], loc='upper left')
plt.show()